10 Python Pandas tips to make data analysis faster
1. Styling
Have you ever complained about the table output looks boring when you do .head() in Jupyter notebooks? Is there a way not to display indexes (especially when there is already an ID column)? There’re ways to fix these issues.
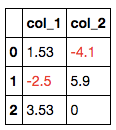
A. Highlight all negative values in a dataframe. (example revised from https://pandas.pydata.org/pandas-docs/stable/user_guide/style.html)
import pandas as pd
def color_negative_red(val):
color = 'red' if val < 0 else 'black'
return 'color: %s' % colordf = pd.DataFrame(dict(col_1=[1.53,-2.5,3.53],
col_2=[-4.1,5.9,0])
)
df.style.applymap(color_negative_red)
B. Hide the index. Try df.head().style.hide_index()!
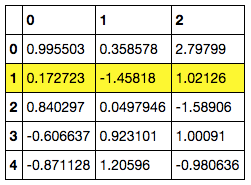
C. Add hovering effects. (example revised from https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.io.formats.style.Styler.set_table_styles.html)
df = pd.DataFrame(np.random.randn(5, 3))
df.style.set_table_styles(
[{'selector': 'tr:hover',
'props': [('background-color', 'yellow')]}]
)
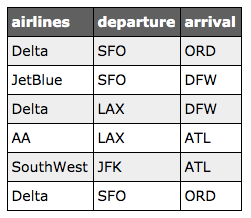
D. More CSS styles. You can use CSS to change the appearance of the table.
df = pd.DataFrame(
dict(departure=['SFO', 'SFO', 'LAX', 'LAX', 'JFK', 'SFO'],
arrival=['ORD', 'DFW', 'DFW', 'ATL', 'ATL', 'ORD'],
airlines=['Delta','JetBlue','Delta',’AA','SouthWest',
'Delta']),
columns=['airlines', 'departure','arrival'])
df.style.set_table_styles(
[{'selector': 'tr:nth-of-type(odd)',
'props': [('background', '#eee')]},
{'selector': 'tr:nth-of-type(even)',
'props': [('background', 'white')]},
{'selector': 'th',
'props': [('background', '#606060'),
('color', 'white'),
('font-family', 'verdana')]},
{'selector': 'td',
'props': [('font-family', 'verdana')]},
]
).hide_index()
2. Pandas options
The reader may have experienced the following issues when using .head(n) to check the dataframe:
(1) There’re too many columns / rows in the dataframe and some columns / rows in the middle are omitted.
(2) Columns containing long texts get truncated.
(3) Columns containing floats display too many / too few digits.
One can set
import pandas as pd
pd.options.display.max_columns = 50 # None -> No Restrictions
pd.options.display.max_rows = 200 # None -> Be careful with this
pd.options.display.max_colwidth = 100
pd.options.display.precision = 3
to solve these issues.
3. Group by with multiple aggregations
In SQL we can do aggregations like
SELECT A, B, max(A), avg(A), sum(B), min(B), count(*)
FROM table
GROUP BY A, B
In Pandas it can be done with .groupby() and .agg():
import pandas as pd
import numpy as np
df = pd.DataFrame(dict(A=['coke', 'sprite', 'coke', 'sprite',
'sprite', 'coke', 'coke'],
B=['alpha','gamma', 'alpha', 'beta',
'gamma', 'beta', 'beta'],
col_1=[1,2,3,4,5,6,7],
col_2=[1,6,2,4,7,9,3]))
tbl = df.groupby(['A','B']).agg({'col_1': ['max', np.mean],
'col_2': ['sum','min','count']})
# 'count' will always be the count for number of rows in each group.
And the result will look like this:
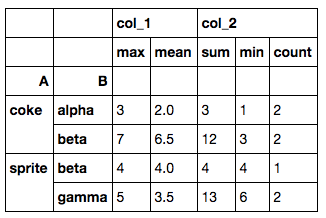
Both the rows and columns are multi-indexed. A quick solution to change it to a dataframe without multi-indices is
tbl = tbl.reset_index()
tbl.columns = ['A', 'B', 'col_1_max', 'col_2_sum', 'col_2_min', 'count']
If you would like to have the column renaming process automated, you can do tbl.columns.get_level_values(0) and tbl.columns.get_level_values(1) to extract the indices in each level and combine them.
4. Column slicing
Some of you might be familiar with this already, but I still find it very useful when handling a dataframe with a ton of columns.
df.iloc[:,2:5].head() # select the 2nd to the 4th column
df.loc[:,'column_x':].head()
# select all columns starting from 'column_x'
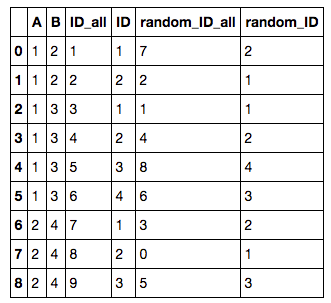
5. Add row ID / random row ID to each group
To add a row ID / random row ID for each group by A, B, one can first append an ID / random ID to all rows:
import numpy as np
# df: target dataframe np.random.seed(0) # set random seed
df['random_ID_all'] = np.random.permutation(df.shape[0])
df['ID_all'] = [i for i in range(1, df.shape[0]+1)]
To add a random ID to each group (by A, B), one can then do
df['ID'] = df.groupby(['A', 'B'])['ID_all'].rank(method='first',ascending=True).astype(int)
df['random_ID'] = df.groupby(['A', 'B'])'random_ID_all'].rank(method='first',ascending=True).astype(int)
to get
6. List all unique values in a group
Sometimes after we performed group by, we’d like to aggregate the values in the target column as a list of unique values instead of max, min, …etc. This is how it’s done.
df = pd.DataFrame(dict(A=['A','A','A','A','A','B','B','B','B'],
B=[1,1,1,2,2,1,1,1,2],
C=['CA','NY','CA','FL','FL',
'WA','FL','NY','WA']))
tbl = df[['A', 'B', 'C']].drop_duplicates()\
.groupby(['A','B'])['C']\
.apply(list)\
.reset_index() # list to string (separated by commas)
tbl['C'] = tbl.apply(lambda x: (','.join([str(s) for s in x['C']])), axis = 1)

If you’d like to save the result, don’t forget to change the separator to anything other than commas.
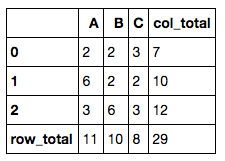
7. Add row total and column total to a numerical dataframe
This is another common data manipulation. All you need is .apply().
df = pd.DataFrame(dict(A=[2,6,3],
B=[2,2,6],
C=[3,2,3]))
df['col_total'] = df.apply(lambda x: x.sum(), axis=1)
df.loc['row_total'] = df.apply(lambda x: x.sum())
8. Check memory usage
**.memory_usage(deep=True)** can be used on Pandas dataframes to see the amount of memory used (in bytes) for each column. It’s useful when building machine learning models which may require a lot memory in training.
9. Cumulative sum
From time to time, cumulative sum is required when you generate some statistical outcomes. Simply do `
df['cumulative_sum'] = df['target_column'].cumsum()` .
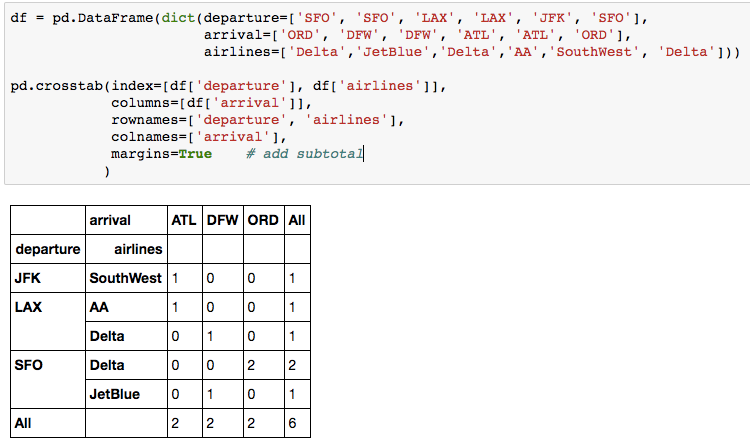
10. Crosstab
When you need to count the frequencies for groups formed by 3+ features, pd.crosstab() can make your life easier.
Thanks for reading! Comment below if you find bugs / better solutions.